Intro
According to U.S. National Cancer Institute, histopathology analysis is description of a tumor based on how abnormal the cancer cells and tissue look under a microscope. Histopathological examination is usually considered the best way to tell if cancer is present . Besides laboriousness of manual identification of tumor tissues, individual pathologist’s previous experience may limit his or her ability to identify certain tumorigenesis given the wide heterogeneity of histologic images. This project aims to train a convolutional neural network (CNN) to aid pathologists to identify presence of tumor tissues from a histologic imagery, as well as to analyze sensitivity, specificity and overall accuracy attained by CNN-based automated histopathological processing.
Description of Technology
The core technology used in this project is convolutional neural network. Specifically, the model was built based on a trained image recognition neural network, VGG16 . This network is comprised of 16 convolutional layers, 14,714,688 parameters. By leveraging the trained VGG16 network, we are expecting to attain better predictive power with relatively small computational burden. Model training was conducted using keras and tensorflow-gpu. The model was trained on Amazon Web Service (AWS p3.2xlarge instance).
Data source and description
The data images and labels were attained from the Histopathologic Cancer Detection challenge presented by Kaggle
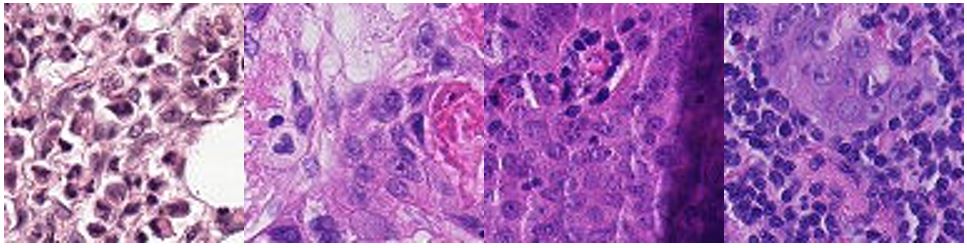
Examples of tumor positive images:
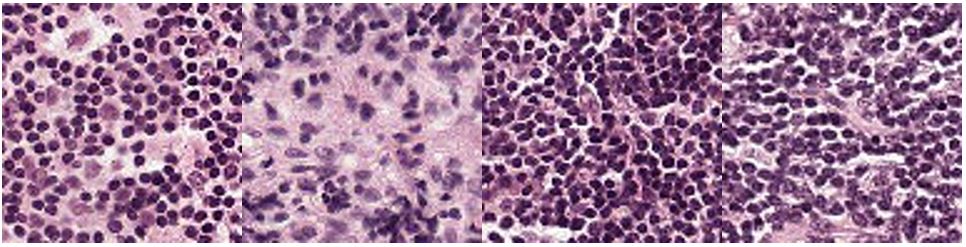
Examples of tumor negative images:
label file. train_labels.csv
| id | label | |
|---|---|---|
| 1 | f38a6374c348f90b587e046aac6079959adf3835 | 0 |
| 2 | c18f2d887b7ae4f6742ee445113fa1aef383ed77 | 1 |
| 3 | 755db6279dae599ebb4d39a9123cce439965282d | 0 |
| 4 | bc3f0c64fb968ff4a8bd33af6971ecae77c75e08 | 0 |
| 5 | 068aba587a4950175d04c680d38943fd488d6a9d | 0 |
| 6 | acfe80838488fae3c89bd21ade75be5c34e66be7 | 0 |
Preprocess data
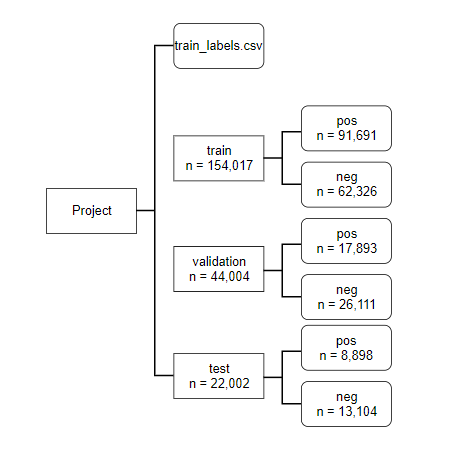
Two steps were taken in data preprocessing. First, we need to separate tumor positive and negative images to different folders (named ‘pos’ and ‘neg’); second, we need to separate out data into training, validation and test sections. Here we employed a 70:20:10 ratio for the three sections.
import os
import pandas as pd
data_dir = r'C:\Users\zkuang\Desktop\histopathologic-cancer-detection'
os.chdir(data_dir)
labels = pd.read_csv('train_labels.csv',header=0)
os.makedirs(r'.\train')
os.makedirs(r'.\neg')
train_cut = int(0.7*labels.shape[0]) # first cutpoint at 0.7
val_cut = int(0.9*labels.shape[0]) # second cutpoint at 0.9
def split_data(start,stop,labels,data_dir,sub_dir):
if not os.path.exists(os.path.join(data_dir,sub_dir,'pos')):
os.makedirs(os.path.join(data_dir,sub_dir,'pos'))
if not os.path.exists(os.path.join(data_dir, sub_dir, 'neg')):
os.makedirs(os.path.join(data_dir, sub_dir, 'neg'))
for i in range(start, stop):
filename = labels.loc[i, 'id']
if labels.loc[i, 'label'] == 1:
try:
os.rename(os.path.join(data_dir, 'raw', filename + '.tif'),
os.path.join(data_dir, sub_dir,'pos', filename + '.tif'))
except FileNotFoundError:
print(filename + ' not found!')
else:
try:
os.rename(os.path.join(data_dir, 'raw', filename + '.tif'),
os.path.join(data_dir, sub_dir,'neg',filename + '.tif'))
except FileNotFoundError:
print(filename + ' not found!')
split_data(0,train_cut,labels,data_dir,'train')# create training data
split_data(train_cut+1,val_cut,labels,data_dir,'validation')# create validation data
split_data(val_cut+1,labels.shape[0],labels,data_dir,'test')# create validation data
Breakdown of training data set-up
Training with pre-trained CNN
import keras
keras.__version__
import logging
import os
Using TensorFlow backend.
#from google.colab import drive
#drive.mount('/content/gdrive')
#os.chdir(r'/content/gdrive/My Drive/FinalProject')
basedir = data_dir
#os.chdir(r'Ubuntu/FinalProject')
#basedir = r'/home/ubuntu/FinalProject'
#logging.basicConfig(filename = r'/content/gdrive/My Drive/FinalProject/test.log',level = logging.INFO)
#logging.basicConfig(filename = r'C:\Users\zkuang\Google Drive\FinalProject\test.log',level = logging.INFO)
logging.basicConfig(filename = os.path.join(basedir,'test2.log'),level = logging.DEBUG)
this is the cell we announce some set numbers with regard to our data
im_size = 96
n_channel = 3
batch_size = 64
Using Keras ImageDataGenerator
Load pretrained model and specify trainable blocks. We allow the last block of VGG16 to be trainable.
from keras.applications import VGG16
conv_base = VGG16(weights='imagenet',
include_top=False,
input_shape=(im_size,im_size,n_channel))
logging.debug(print(conv_base.summary()))
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input_1 (InputLayer) (None, 96, 96, 3) 0
_________________________________________________________________
block1_conv1 (Conv2D) (None, 96, 96, 64) 1792
_________________________________________________________________
block1_conv2 (Conv2D) (None, 96, 96, 64) 36928
_________________________________________________________________
block1_pool (MaxPooling2D) (None, 48, 48, 64) 0
_________________________________________________________________
block2_conv1 (Conv2D) (None, 48, 48, 128) 73856
_________________________________________________________________
block2_conv2 (Conv2D) (None, 48, 48, 128) 147584
_________________________________________________________________
block2_pool (MaxPooling2D) (None, 24, 24, 128) 0
_________________________________________________________________
block3_conv1 (Conv2D) (None, 24, 24, 256) 295168
_________________________________________________________________
block3_conv2 (Conv2D) (None, 24, 24, 256) 590080
_________________________________________________________________
block3_conv3 (Conv2D) (None, 24, 24, 256) 590080
_________________________________________________________________
block3_pool (MaxPooling2D) (None, 12, 12, 256) 0
_________________________________________________________________
block4_conv1 (Conv2D) (None, 12, 12, 512) 1180160
_________________________________________________________________
block4_conv2 (Conv2D) (None, 12, 12, 512) 2359808
_________________________________________________________________
block4_conv3 (Conv2D) (None, 12, 12, 512) 2359808
_________________________________________________________________
block4_pool (MaxPooling2D) (None, 6, 6, 512) 0
_________________________________________________________________
block5_conv1 (Conv2D) (None, 6, 6, 512) 2359808
_________________________________________________________________
block5_conv2 (Conv2D) (None, 6, 6, 512) 2359808
_________________________________________________________________
block5_conv3 (Conv2D) (None, 6, 6, 512) 2359808
_________________________________________________________________
block5_pool (MaxPooling2D) (None, 3, 3, 512) 0
=================================================================
Total params: 14,714,688
Trainable params: 14,714,688
Non-trainable params: 0
_________________________________________________________________
None
This is the part that allows for fine tuning
conv_base.trainable = True
set_trainable = False
for layer in conv_base.layers:
if layer.name == 'block5_conv1':
set_trainable = True
if set_trainable:
layer.trainable = True
else:
layer.trainable = False
Model set up
Added flattening layer and dense network. The last layer has a sigmoid activation function, since our output is binary
from keras import models
from keras import layers
from keras import optimizers
from keras.preprocessing.image import ImageDataGenerator
from keras.models import load_model
import json
model = models.Sequential()
model.add(conv_base) # how you used a trained model
model.add(layers.Flatten())
model.add(layers.Dense(256, activation='relu'))
model.add(layers.Dense(1, activation='sigmoid'))
Set up data generator. Notice that images were standardized before training (rescale = 1./255). Batch size used here is the global batch size 64.
datagen = ImageDataGenerator(rescale=1./255)
train_dir = os.path.join(basedir,'train')
validation_dir = os.path.join(basedir,'validation')
train_generator = datagen.flow_from_directory(
# This is the target directory
train_dir,
# All images will be resized to 150x150
target_size=(im_size, im_size),
batch_size=batch_size,
class_mode='binary')
validation_generator = datagen.flow_from_directory(
validation_dir,
target_size=(im_size, im_size),
batch_size=batch_size,
class_mode='binary')
Found 154017 images belonging to 2 classes.
Found 44004 images belonging to 2 classes.
Model compilation and fitting. We saved our model as an h5 object. This if – else statement made sure that model fitting only takes place when there isn’t a trained model available. This was for the convenience of debugging and model diagnostics. We ran 80 epochs for training and each epoch took 100 steps.
model.compile(loss='binary_crossentropy',
optimizer=optimizers.RMSprop(lr=1e-5),
metrics=['acc'])
model.summary()
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
vgg16 (Model) (None, 3, 3, 512) 14714688
_________________________________________________________________
flatten_1 (Flatten) (None, 4608) 0
_________________________________________________________________
dense_1 (Dense) (None, 256) 1179904
_________________________________________________________________
dense_2 (Dense) (None, 1) 257
=================================================================
Total params: 15,894,849
Trainable params: 8,259,585
Non-trainable params: 7,635,264
_________________________________________________________________
Model run
if os.path.isfile(os.path.join(basedir,'hist_path.h5')): # only train the model when it's not already existent
model = load_model(os.path.join(basedir,'hist_path.h5'))
with open('model_call_back.json', 'r') as f:
history = json.load(f)
else:
history = model.fit_generator(
train_generator,
steps_per_epoch=100,
epochs=120,
validation_data=validation_generator,
validation_steps=50)
history = history.history
model.save(os.path.join(basedir,'hist_path.h5'))
with open('model_call_back.json', 'w') as f:
json.dump(history, f)
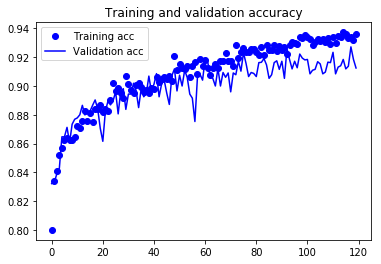
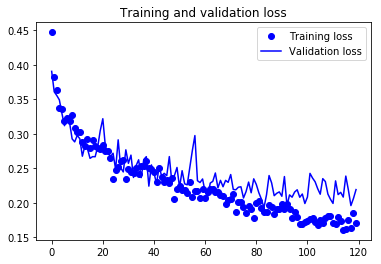
Plotting cost and accuracy. This was the part we conducted internal model diagnostics. Loss and accuracy in training samples and validation samples were plotted against epochs.
import matplotlib.pyplot as plt
%matplotlib inline
acc = history['acc']
val_acc = history['val_acc']
loss = history['loss']
val_loss = history['val_loss']
epochs = range(len(acc))
plt.plot(epochs, acc, 'bo', label='Training acc')
plt.plot(epochs, val_acc, 'b', label='Validation acc')
plt.title('Training and validation accuracy')
plt.legend()
plt.savefig('accuracy.png')
plt.figure()
plt.plot(epochs, loss, 'bo', label='Training loss')
plt.plot(epochs, val_loss, 'b', label='Validation loss')
plt.title('Training and validation loss')
plt.legend()
plt.show()
plt.savefig('loss.png')
<Figure size 432x288 with 0 Axes>
Investigate accuracy, sensitivity and specificity
The verification steps yielded pictures as follow, indicating proper data maneuvering for the test step.
from PIL import Image
import numpy as np
test_dir = os.path.join(basedir,'test')
test_n_pos = len(os.listdir(os.path.join(test_dir,'pos')))
pos_features = np.zeros(shape=(test_n_pos, im_size, im_size, 3))
i = 0
for file in os.listdir(os.path.join(test_dir,'pos')):
pos_features[i] = np.divide(np.array(Image.open(os.path.join(test_dir,'pos',file))),255)
i+=1
test_n_neg = len(os.listdir(os.path.join(test_dir,'neg')))
neg_features = np.zeros(shape=(test_n_neg, im_size, im_size, 3))
i = 0
for file in os.listdir(os.path.join(test_dir,'neg')):
neg_features[i] = np.divide(np.array(Image.open(os.path.join(test_dir,'neg',file))),255)
i+=1
last_tif = (pos_features[1]*255).astype(int)
plt.imshow(last_tif)
last_tif = (neg_features[1]*255).astype(int)
plt.imshow(last_tif)
if os.path.isfile(os.path.join(basedir,'y_pos.txt')):
y_pos = np.loadtxt("y_pos.txt")
y_neg = np.loadtxt("y_neg.txt")
else:
y_pos = model.predict(pos_features)
y_pos = (y_pos>0.5) *1
np.savetxt("y_pos.txt", y_pos, delimiter=",")
y_neg = model.predict(neg_features)
y_neg = (y_neg>0.5) *1
np.savetxt("y_neg.txt", y_neg, delimiter=",")
print('true positive = ')
print(sum(y_pos))
print('false negative = ')
print(sum(1-y_pos))
print('true negative = ')
print(sum(1-y_neg))
print('false positive = ')
print(sum(y_neg))
true positive =
7987.0
false negative =
911.0
true negative =
12212.0
false positive =
892.0
print('sensitivity =')
print(sum(y_pos)/len(y_pos))
print('specificity =')
print(sum(1-y_neg)/len(y_neg))
print('overral accuracy = ')
print((sum(y_pos) + sum(1-y_neg)) /
(len(y_pos)+len(y_neg)))
sensitivity =
0.8976174421218251
specificity =
0.931929181929182
overral accuracy =
0.9180529042814289
The model attained sensitivity of 0.899, specificity of 0.932. The overall accuracy of our model is 0.918, which is comparable to state-of-art supervised machine learning histopathological image recognition results. Given our images are of lower resolution than most studies use (often ultrahigh resolution imageries with more than 1 million pixels ), the results should be deemed particularly remarkable. It is no surprising that we attained better specificity than sensitivity, given our data is slightly skewed towards negative cases.
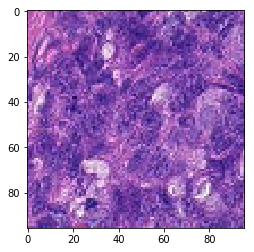
# visualization of misclassified images
print('Mis-classified positive images')
j = 0
for i in range(len(y_pos)):
if y_pos[i] == 0:
tif = (pos_features[i]*255).astype(int)
plt.figure()
plt.imshow(tif)
j += 1
if j == 5:
break
Mis-classified positive images
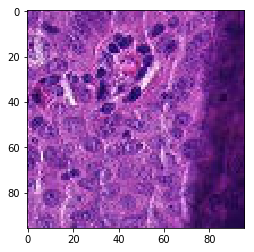
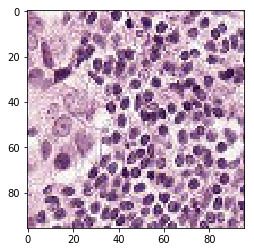
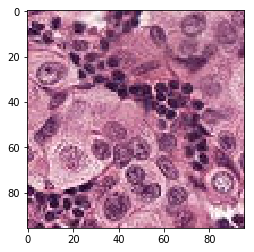
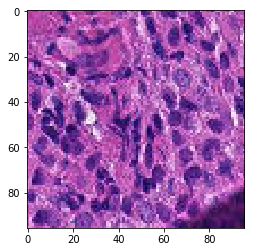
print('Mis-classified negative images')
j = 0
for i in range(len(y_neg)):
if y_neg[i] == 1:
tif = (neg_features[i]*255).astype(int)
plt.figure()
plt.imshow(tif)
j += 1
if j == 5:
break
Mis-classified negative images
it’s is difficult to intuitively elucidate why some of the images are mis-classified (such as shown in the previous section). One can conjecture that less stained imageries such as 1) and 5) in misclassified negative images are more likely to be considered negative by our algorithm, versus slides with high internal heterogeneity as seen in misclassified positive images are more likely to be falsely classified as cancerous. Therefore, it will be interesting to bring together human pathologist and data scientist to better illuminate the strengths and weakness of CNN-based histopathological analyses.
Due to the limitations of computational power, we didn’t try hyperparameter tuning for our model. It was conjectured a deeper convolutional neural net and relatively large epoch size (n= 120) should suffice to attain a reasonable approximation of the optimal model. Indeed, from the internal diagnostics we saw valuation accuracy tapering off after roughly 80 epochs. Whether or not fine-tuning learning rate can improve model performance remained to be examined. Similarly, we didn’t experiment with setting different VGG16 blocks trainable, the effect of which also remained to be examined.